The OptForce procedure identifies the minimal set of genetic interventions that shape the metabolism of a microorganism so as it guarantees a pre-specified biochemical product target yield. The procedure is designed to make use of flux measurements, whenever available, for the wild-type strain. OptForce works by comparing the allowable range of flux variability for a wild-type strain and the overproducing network. By comparing fluxes one-at-a-time (see Figure 1) we first identify reactions whose flux must increase (MUSTU), decrease (MUSTL) or completely shut-off (MUSTX) consistent with the overproduction target. For fluxes with overlapping ranges in the overproducing network, we compare sums (or differences) of two fluxes at a time (see Figure 2 and Non-overlapping sums imply that the flux of either one reaction OR the other MUST increase. These reaction pairs are appended within the MUSTUU set. We use a similar classification procedure to identify the pairs of reactions that populate MUSTUL and MUSTLL sets.
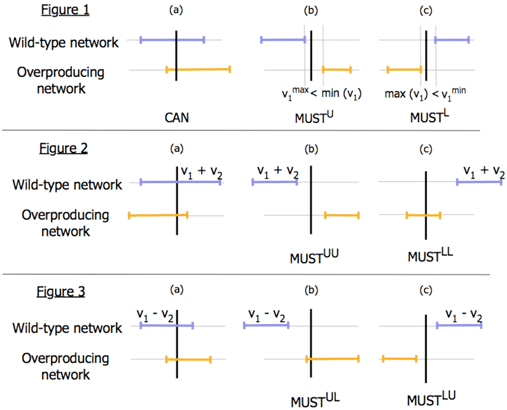
Note that not all reactions in the MUST sets need to be actively engineered for overproduction. Typically, only a subset needs to be engineer and propagate this effect to all other members of the MUST sets by virtue of stoichiometry and other constraints. We use a bilevel optimization framework to identify the minimal set of engineering interventions that FORCE the wild-type network phenotype towards the overproducing network.
Related Publications:
Ranganathan. S, Suthers. P. F., and Maranas. C. D. (2010), "OptForce: An optimization procedure for identifying all genetic manipulations leading to targeted overproductions", PLoS Computational Biology 6(4): e1000744. doi:10.1371/journal.pcbi.1000744
Ranganathan. S, and Maranas. C. D. (2010), "Microbial 1-butanol production: Identification of non-native production routes and in silico engineering interventions", Biotechnology Journal 5(7) 716. doi:10.1002/biot.201000171
Xu .P, Ranganathan .S, Fowler. Z. L., Maranas. C. D., Koffas. M. A. (2011), "Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA", Metabolic Engineering doi:10.1016/j.ymben.2011.06.008
Chowdhury A., Zomorrodi A.R., Maranas C.D. (2015), "Bilevel optimization techniques in computational strain design", Computers and Chemical Engineering, 72:363-372.
The code is freely available for academic users. Before downloading our software, we ask for your name and institution so we may better design future versions of our software. Every consideration has been made during the design, writing, and posting of this code to ensure the program works properly.
